Performs Gauss-Newton weighted least squares computation. More...
#include <wls.h>
Public Member Functions | |
| WLS (int size=0) | |
| void | clear () |
| void | add_prior (Precision val) |
| template<class B2 > | |
| void | add_prior (const Vector< Size, Precision, B2 > &v) |
| template<class B2 > | |
| void | add_prior (const Matrix< Size, Size, Precision, B2 > &m) |
| template<class B2 > | |
| void | add_mJ (Precision m, const Vector< Size, Precision, B2 > &J, Precision weight=1) |
| template<int N, class B1 , class B2 , class B3 > | |
| void | add_mJ (const Vector< N, Precision, B1 > &m, const Matrix< Size, N, Precision, B2 > &J, const Matrix< N, N, Precision, B3 > &invcov) |
| template<int N, class B1 , class B2 , class B3 > | |
| void | add_mJ_rows (const Vector< N, Precision, B1 > &m, const Matrix< N, Size, Precision, B2 > &J, const Matrix< N, N, Precision, B3 > &invcov) |
| template<int N, typename B1 > | |
| void | add_sparse_mJ (const Precision m, const Vector< N, Precision, B1 > &J1, const int index1, const Precision weight=1) |
| template<int N, int S1, class P1 , class P2 , class P3 , class B1 , class B2 , class B3 > | |
| void | add_sparse_mJ_rows (const Vector< N, P1, B1 > &m, const Matrix< N, S1, P2, B2 > &J1, const int index1, const Matrix< N, N, P3, B3 > &invcov) |
| template<int N, int S1, int S2, class B1 , class B2 , class B3 , class B4 > | |
| void | add_sparse_mJ_rows (const Vector< N, Precision, B1 > &m, const Matrix< N, S1, Precision, B2 > &J1, const int index1, const Matrix< N, S2, Precision, B3 > &J2, const int index2, const Matrix< N, N, Precision, B4 > &invcov) |
| void | compute () |
| void | operator+= (const WLS &meas) |
| Matrix< Size, Size, Precision > & | get_C_inv () |
|
const Matrix< Size, Size, Precision > & | get_C_inv () const |
| Vector< Size, Precision > & | get_mu () |
| const Vector< Size, Precision > & | get_mu () const |
| Vector< Size, Precision > & | get_vector () |
| const Vector< Size, Precision > & | get_vector () const |
| Decomposition< Size, Precision > & | get_decomposition () |
|
const Decomposition< Size, Precision > & | get_decomposition () const |
Detailed Description
template<int Size = Dynamic, class Precision = DefaultPrecision, template< int DecompSize, class DecompPrecision > class Decomposition = Cholesky>
class TooN::WLS< Size, Precision, Decomposition >
Performs Gauss-Newton weighted least squares computation.
- Parameters:
-
Size The number of dimensions in the system Precision The numerical precision used (double, float etc) Decomposition The class used to invert the inverse Covariance matrix (must have one integer size and one typename precision template arguments) this is Cholesky by default, but could also be SQSVD
Member Function Documentation
| void add_prior | ( | Precision | val | ) |
Applies a constant regularisation term.
Equates to a prior that says all the parameters are zero with 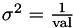 .
.
- Parameters:
-
val The strength of the prior
| void add_prior | ( | const Vector< Size, Precision, B2 > & | v | ) |
Applies a regularisation term with a different strength for each parameter value.
Equates to a prior that says all the parameters are zero with 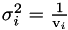 .
.
- Parameters:
-
v The vector of priors
| void add_prior | ( | const Matrix< Size, Size, Precision, B2 > & | m | ) |
Applies a whole-matrix regularisation term.
This is the same as adding the 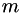 to the inverse covariance matrix.
to the inverse covariance matrix.
- Parameters:
-
m The inverse covariance matrix to add
| void add_mJ | ( | Precision | m, |
| const Vector< Size, Precision, B2 > & | J, | ||
| Precision | weight = 1 |
||
| ) |
Add a single measurement.
- Parameters:
-
m The value of the measurement J The Jacobian for the measurement 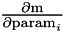
weight The inverse variance of the measurement (default = 1)
| void add_mJ | ( | const Vector< N, Precision, B1 > & | m, |
| const Matrix< Size, N, Precision, B2 > & | J, | ||
| const Matrix< N, N, Precision, B3 > & | invcov | ||
| ) |
Add multiple measurements at once (much more efficiently)
- Parameters:
-
m The measurements to add J The Jacobian matrix 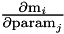
invcov The inverse covariance of the measurement values
| void add_mJ_rows | ( | const Vector< N, Precision, B1 > & | m, |
| const Matrix< N, Size, Precision, B2 > & | J, | ||
| const Matrix< N, N, Precision, B3 > & | invcov | ||
| ) |
Add multiple measurements at once (much more efficiently)
- Parameters:
-
m The measurements to add J The Jacobian matrix 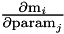
invcov The inverse covariance of the measurement values
| void add_sparse_mJ | ( | const Precision | m, |
| const Vector< N, Precision, B1 > & | J1, | ||
| const int | index1, | ||
| const Precision | weight = 1 |
||
| ) |
Add a single measurement at once with a sparse Jacobian (much, much more efficiently)
- Parameters:
-
m The measurements to add J1 The first block of the Jacobian matrix 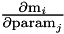
index1 starting index for the first block invcov The inverse covariance of the measurement values
| void add_sparse_mJ_rows | ( | const Vector< N, P1, B1 > & | m, |
| const Matrix< N, S1, P2, B2 > & | J1, | ||
| const int | index1, | ||
| const Matrix< N, N, P3, B3 > & | invcov | ||
| ) |
Add multiple measurements at once with a sparse Jacobian (much, much more efficiently)
- Parameters:
-
m The measurements to add J1 The first block of the Jacobian matrix 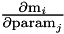
index1 starting index for the first block invcov The inverse covariance of the measurement values
| void add_sparse_mJ_rows | ( | const Vector< N, Precision, B1 > & | m, |
| const Matrix< N, S1, Precision, B2 > & | J1, | ||
| const int | index1, | ||
| const Matrix< N, S2, Precision, B3 > & | J2, | ||
| const int | index2, | ||
| const Matrix< N, N, Precision, B4 > & | invcov | ||
| ) |
Add multiple measurements at once with a sparse Jacobian (much, much more efficiently)
- Parameters:
-
m The measurements to add J1 The first block of the Jacobian matrix 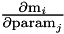
index1 starting index for the first block J2 The second block of the Jacobian matrix 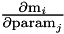
index2 starting index for the second block invcov The inverse covariance of the measurement values
 1.7.4
1.7.4